Over the last 5 decades we have seen a dramatic increase in the incidence of several immune mediated conditions, including IBD. This increase has been particularly prominent in the western world. However, interestingly, many developing countries that are adapting to a “western” life style are now also reporting a rapid increase in patients diagnosed with IBD. In the absence of much evidence for changes to the human genome in the same short time, we hypothesise that changes in our environment are key drivers of complex disease development.
Epigenetic mechanisms provide the interface between the genome and the environment, translating the effects of environmental change into a change in phenotype. Specifically, epigenetics can be defined as changes in phenotype, i.e. gene expression, caused by mechanisms other than changes in the underlying DNA sequence. The main epigenetic mechanisms known to be operative in humans include post-transcriptional histone modification, expression of non-coding RNAs (e.g. siRNA, miRNA) and DNA methylation. The latter is amongst the most extensively studied and has been shown to be implicated in several fundamental biological processes, e.g. genomic imprinting, X-chromosome inactivation and regulating cell type specific gene expression/cellular function. One of the key research areas in our group is to investigate the role of epigenetic mechanisms, particularly DNA methylation, in intestinal health and IBD in particular.
We have previously shown that DNA methylation profiles are highly cell type specific and that patient derived organoids (human mini guts) retain organ, cell type and even disease specific information. Our large clinical cohort provides us with access to tissue from children diagnosed with IBD, as well as age matched healthy controls. As a result, organoids can now be used to study epigenetic mechanisms and develop novel treatments (Figure 2).
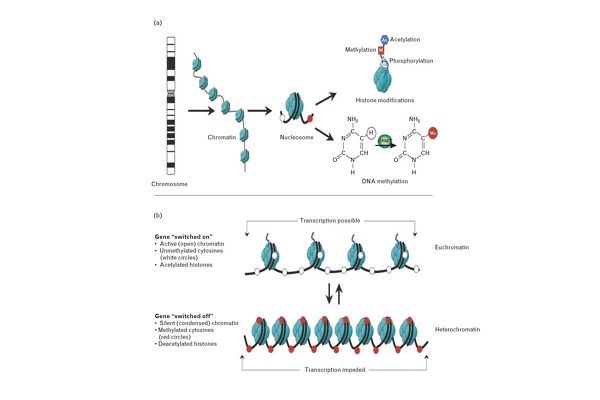
Figure 1: Epigenetic modifications, their effect on chromatin state and gene transcription. (a) DNA wrapped around histone octamers forming nucleosomes, which represent the principal component of chromatin. Reversible and site-specific posttranslational histone modifications occur at multiple sites through acetylation, methylation and phosphorylation. DNA methylation occurs at 50 position of cytosine residues in a reaction catalysed by DNA methyltransferases (DNMTs). (b) DNA methylation and h
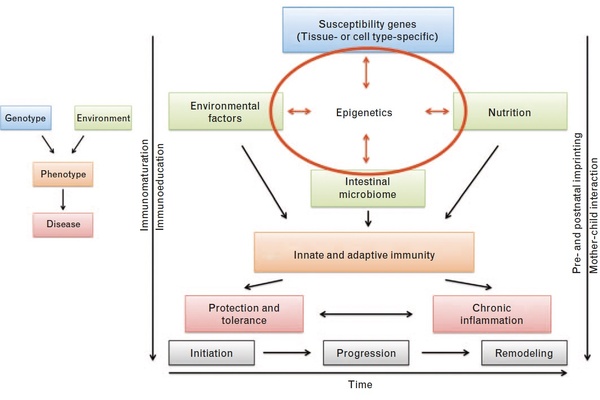
Figure 2: Revised model of inflammatory bowel disease (IBD) pathogenesis. (a) Traditional simplistic paradigm of IBD pathogenesis. (b) Revised, more complex model of IBD pathogenesis in which epigenetics plays a central role in mediating effects of the key cornerstones: genetic predisposition, environmental factors, nutrition and the intestinal microbiome.
Jenke and Zilbauer, Current Opinion in Immunology, 2012